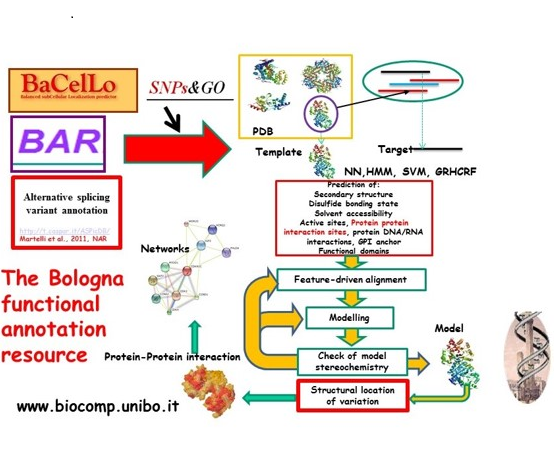
Research themes
Development of algorithms for structural and functional annotation of proteins and their variants. The main problem of the “genomic era” is the ever-increasing mole of data collected at different levels of cell complexity. It is then fundamental to extract information useful to a better understanding of the rules governing the hierarchical organization of the biological complexity and to explaining of the emergence of physiological processes.
The non-redundant information extracted from data bases is analyzed with algorithms, routinely based on statistics, probability theory and machine learning approaches. This allows inferring structural and functional features of molecules poorly characterized from the experimental point of view. The outcome is an algorithm, usually called “predictor”, that is able to associate the probability for a given feature to be associated or not to the molecule under investigation (http://www.biocomp.unibo.it/predictors.html).
Protein-protein interactions in different organisms and their integration in models of Systems Biology. The key for understanding biological complexity is the structural and functional interaction among molecules into the different sub-cellular compartments.
Protein-protein interaction is at the basis of metabolic pathways and biological processes. The genomic variation of one or more components can lead to a loss of functionality of the whole system. A technique, called enrichment, can be used to elucidate in a statistically validated way whether different variations affect one or more biological processes (http://net-ge.biocomp.unibo.it/)
Relationship between genotype and phenotype. Integration of structural, functional and genomic data for dissecting the relationship between genotype and phenotype.
It is possible to draft hypotheses on the relationship between human genotype and phenotype, in particular in relation to diseases, by statistically validating the enrichment of structural and functional annotations of the proteins associated to the same phenotype. This study allows an integrated view of the complex interplay between genotype and phenotype (http://edgar.biocomp.unibo.it/phenpath)
(http://edgar.biocomp.unibo.it/gene_disease_db/)
Development of molecular models for complex phenotypes. The onset of diseases can depend on different molecular determinants, often specific for each individual.
The integrated analysis of inherited and somatic variations associated to different diseases, and in particular to cancers, prompts for the discrimination of molecular mechanisms that can help researchers in designing new experiments. NGS data analysis in the perspective of functional genomics allows developing models able to associate a set of mutations with the probabilistic risk for the disease onset.
Aspects of the structure-function relationship in membrane ATPases. Relationship between the number of protons per ATP molecule translocated across the membrane and the number of transmembrane subunits in the F-type ATPases.
The F-type ATPases can be viewed as nanoscopic rotational motors, in which a transmembrane rotor is driven by a proton gradient relative to an extramembrane stator which synthesizes ATP. We have set up a method for the experimental determination of the number of translocated protons for each synthetized ATP molecule. Recently, we have obtained evidence that such number can be variable, depending on the energetic state of the cell.