Helicobacter pylori is an important human pathogen recognized by the World Health Organization as a class I carcinogen.
The ability of this Gram-negative bacterium to colonize the hostile stomach niche and to establish a persistent infection depends on the coordinated expression of virulence factors that allow the pathogen to adapt to adverse environmental conditions and to counteract the defense mechanisms of the host. The research carried out by our group combines molecular biology, biochemical, and genetic methodologies to study the molecular mechanisms adopted by two transcriptional repressors involved in the heat-shock response, HrcA and HspR, of a response regulator essential for the viability of the bacterium called HP1043 involved in the regulation of crucial cellular processes, and of a small regulatory RNA involved in bacterial motility, called CncR1.
Research themes of the group
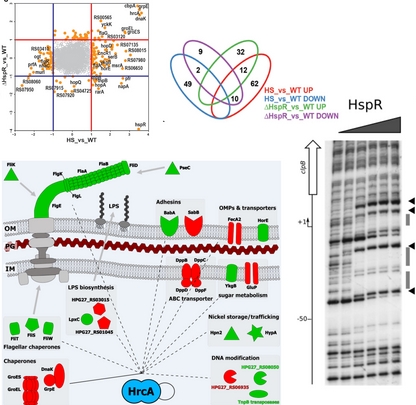
Transcriptional Control in Response to Heat-shock:
The heat-shock response in the human pathogen H. pylori is governed by two transcriptional repressors, HrcA and HspR. Under normal growth conditions, these regulators maintain the repression of the main heat-shock genes and, following thermal shock, allow a rapid and transient accumulation of the regulated transcripts. Our group studies the molecular mechanisms exerted by each regulator and their interactions to control the transcription of genes in response to temperature fluctuations.
Transcriptional Control by the Essential Regulator HP1043:
The HP1043 transcriptional regulator is encoded by a gene essential for the vitality of the bacterium. The inability to generate a deletion mutant for the hp1043 gene or to modulate the protein levels of the regulator in vivo has so far precluded the understanding of its function. Through chromatin immunoprecipitation assays (ChIP), we identified 37 promoters bound in vivo by HP1043 at the genomic level, which allowed the identification of the DNA-binding consensus sequences in vitro. We study the molecular mechanisms of in vitro transcription activation by identifying the determinants of DNA binding specificity and the effects on its ability to activate transcription. In parallel, we will generate an hp1043 conditional mutant that will allow defining the impact of HP1043 gene silencing on the H. pylori transcriptome.
Moreover, being HP1043 a promising antibacterial target due to its central regulatory role, we are carrying on in silico protein-DNA docking and molecular dynamics studies for the virtual screening of molecule libraries aiming at the identification of compounds capable of binding and interfering with the activity of the HP1043 protein and, consequently, with the viability of H. pylori.
Definition of the regulatory role and mechanism of action of non-coding small RNAs in H. pylori
Recent studies demonstrated that H. pylori encodes massive amounts of non-coding transcripts that are expected to regulate gene expression through different mechanisms posttranscriptionally.
By combining molecular biology techniques (like generation of mutants, RNA-RNA interaction assays, and RNA structure probing) with functional genomics approaches (RNA-seq, comparative proteomics, and others), our group is characterizing the regulatory role and the mechanism of action of some H. pylori non-coding RNA. For example, to understand the regulatory role of CncR1, a non-coding RNA whose transcription is regulated by HP1043, we have set up a recently developed functional genomics approach called MAPS (MS2 affinity purification coupled with RNA sequencing) to define CncR1 interactome.